Nanobiotics: model predicts how nanoparticles interact with proteins
Nano-engineered drugs that stop harmful bacteria and viruses could be on the horizon.

Nano-engineered drugs that stop harmful bacteria and viruses could be on the horizon.
U-M EXPERTS:
Originally written by Kate McAlpine and published by Michigan Engineering.
With antibiotic-resistant infections on the rise and a continually morphing pandemic virus, it’s easy to see why researchers want to be able to design engineered nanoparticles that can shut down these infections. A new machine learning model that predicts interactions between nanoparticles and proteins, developed at the University of Michigan, brings us a step closer to that reality.
“We have reimagined nanoparticles to be more than mere drug delivery vehicles. We consider them to be active drugs in and of themselves,” said J. Scott VanEpps, an assistant professor of emergency medicine and an author of the study in Nature Computational Science.
Discovering drugs is a slow and unpredictable process, which is why so many antibiotics are variations on a previous drug. Drug developers would like to design medicines that can attack bacteria and viruses in ways that they choose, taking advantage of the “lock-and-key” mechanisms that dominate interactions between biological molecules. But it was unclear how to transition from the abstract idea of using nanoparticles to disrupt infections to practical implementation of the concept.
“By applying mathematical methods to protein-protein interactions, we have streamlined the design of nanoparticles that mimic one of the proteins in these pairs,” said Nicholas Kotov, the Irving Langmuir Distinguished University Professor of Chemical Sciences and Engineering and corresponding author of the study.
“Nanoparticles are more stable than biomolecules and can lead to entirely new classes of antibacterial and antiviral agents.”
The new machine learning algorithm compares nanoparticles to proteins using three different ways to describe them. While the first was a conventional chemical description, the two that concerned structure turned out to be most important for making predictions about whether a nanoparticle would be a lock-and-key match with a specific protein.
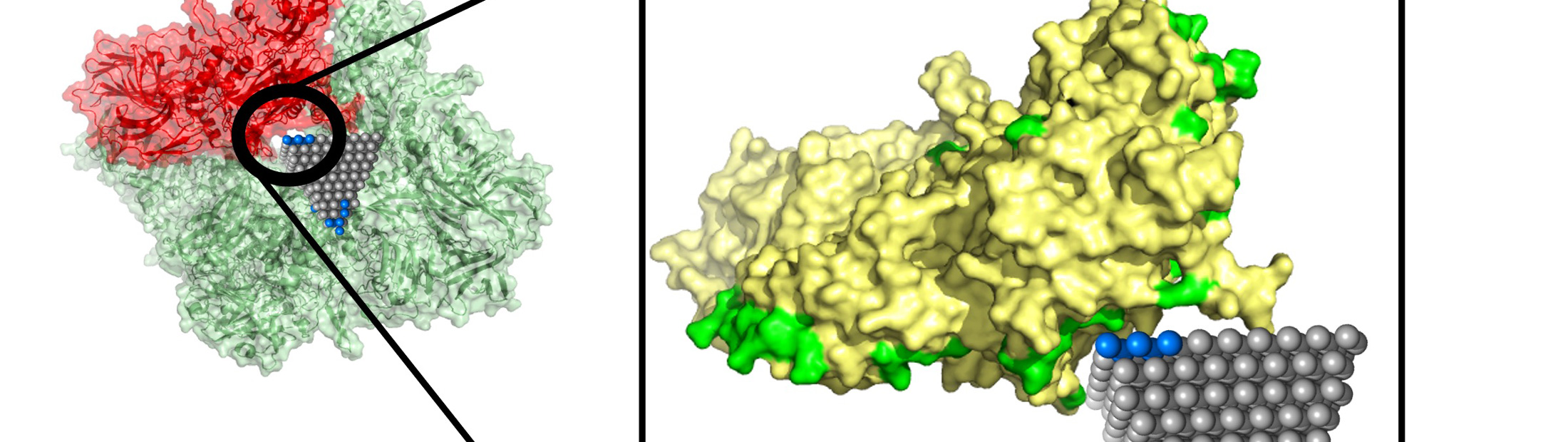
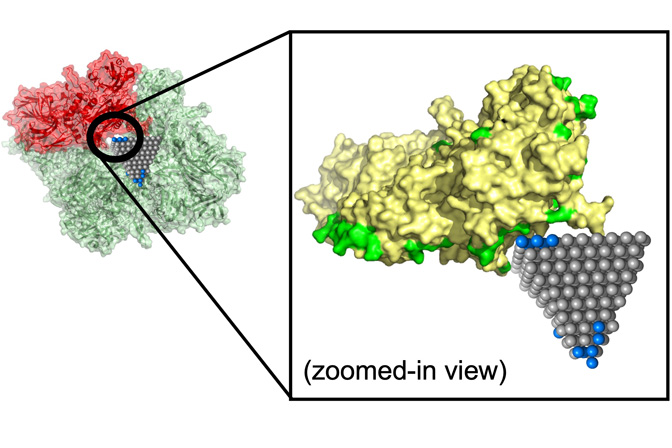
Between them, these two structural descriptions captured the protein’s complex surface and how it might reconfigure itself to enable lock-and-key fits. This includes pockets that a nanoparticle could fit into, along with the size such a nanoparticle would need to be. The descriptions also included chirality, a clockwise or counterclockwise twist that is important for predicting how a protein and nanoparticle will lock in.
“There are many proteins outside and inside bacteria that we can target. We can use this model as a first screening to discover which nanoparticles will bind with which proteins,” said Emine Sumeyra Turali Emre, a postdoctoral researcher in chemical engineering and co-first author of the paper, along with Minjeong Cha, a PhD student in materials science and engineering.
Emre and Cha explained that researchers could follow up on matches identified by their algorithm with more detailed simulations and experiments. One such match could stop the spread of MRSA, a common antibiotic-resistant strain, using zinc oxide nanopyramids that block metabolic enzymes in the bacteria.
“Machine learning algorithms like ours will provide a design tool for nanoparticles that can be used in many biological processes. Inhibition of the virus that causes COVID-19 is one good example,” said Cha. “We can use this algorithm to efficiently design nanoparticles that have broad-spectrum antiviral activity against all variants.”
This breakthrough was enabled by the Blue Sky Initiative at the University of Michigan College of Engineering. It provided $1.5 million to support the interdisciplinary team carrying out the fundamental exploration of whether a machine learning approach could be effective when data on the biological activity of nanoparticles is so sparse.
“The core of the Blue Sky idea is exactly what this work covers: finding a way to represent proteins and nanoparticles in a unified approach to understand and design new classes of drugs that have multiple ways of working against bacteria,” said Angela Violi, an Arthur F. Thurnau Professor, a professor of mechanical engineering and leader of the nanobiotics Blue Sky project.
Emre led the building of a database of interactions between proteins that could help to predict nanoparticle and protein interaction. Cha then identified structural descriptors that would serve equally well for nanoparticles and proteins, working with collaborators at the University of Southern California, Los Angeles to develop a machine learning algorithm that combed through the database and used the patterns it found to predict how proteins and nanoparticles would interact with one another. Finally, the team compared these predictions for lock-and-key matches with the results from experiments and detailed simulations, finding that they closely matched.
Additional collaborators on the project include Ji-Young Kim, a postdoctoral researcher in chemical engineering at U-M, who helped calculate chirality in the proteins and nanoparticles. Paul Bogdan and Xiongye Xiao, a professor and PhD student, respectively, in electrical and computer engineering at USC contributed to the graph theory descriptors. Cha then worked with them to design and train the neural network, comparing different machine learning models. All authors helped analyze the data.
In addition to the University of Michigan College of Engineering’s Blue Sky Initiative, this study was funded by the Office of Naval Research through grants N000141812876 and HQ00342010033, and the Air Force Office of Scientific Research grant FA9550-20-1-0265.
Kotov is also the Joseph B. and Florence V. Cejka Professor of Engineering and a professor of chemical engineering, materials science and engineering and macromolecular science and engineering. Violi is also a professor of chemical engineering and biophysics. VanEpps is also a professor of macromolecular science and engineering.
“Universal descriptors to predict interactions of inorganic nanoparticles with proteins.”