
Precision health and advanced communications: €9M ($10M) for bio-inspired nanoparticles on demand
Advanced microscopy techniques and AI models will help design complex nanoparticles for specific biological targets with less trial and error.

Advanced microscopy techniques and AI models will help design complex nanoparticles for specific biological targets with less trial and error.
Written by Derek Smith
Customized nanoparticles with complex shapes could help fight disease or transmit information, and the European Research Council is investing €9.3 million (~$10.1 million) in a research team co-led by the University of Michigan to speed up nanoparticle design for these applications.
The team is also co-led by the Centre for Cooperative Research in Biomaterials-CIC biomaGUNE and the University of Vigo in Spain, as well as the University of Antwerp in Belgium. They aim to design a machine-learning model that can predict which nanoparticle structures will bind to specific biomolecules, such as proteins on the cell walls of pathogens.
Selective binding to various proteins and lipids (components of cell membranes) is directly useful in detecting and treating diseases. For instance, nanoparticles designed to twist like their protein targets have already been used to detect molecular signs of Parkinson’s disease and lung cancer in laboratory studies.
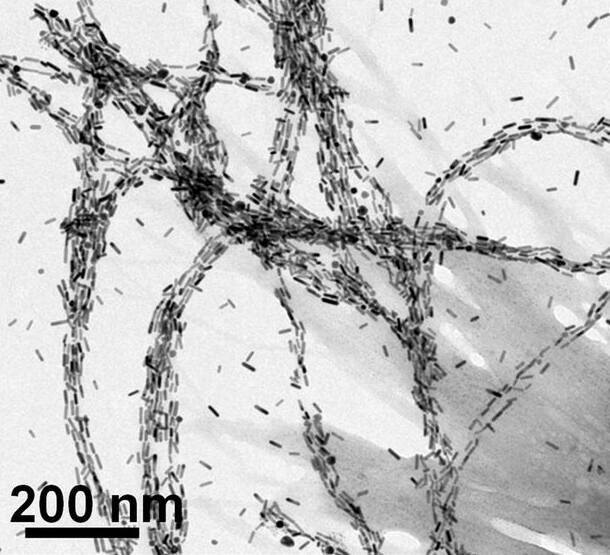
The direction of the nanoparticles’ twists, or their chirality, not only determines how strongly they bind with certain proteins, but it also impacts whether they absorb or twist specific colors and orientations of light. Researchers can detect the change to confirm the presence of the target protein. Light can also be used to selectively heat the protein, killing only the harmful cells attached to the nanoparticles.
Designing chiral nanoparticles that can bind to specific biological targets remains a challenge, however.
“We have to understand when the nanoparticles form strong complexes with specific proteins like a lock and key, but predicting those complexes is hard,” said Nick Kotov, the Irving Langmuir Distinguished University Professor of Chemical Sciences and Engineering at the University of Michigan and a co-principal investigator on the project.
“Many biomolecules in the body could interact with simple, spherical nanoparticles, but these interactions are nonspecific. We need to make nanoparticles with more complex shapes across different scales that recognize and bind to biological targets with high specificity.”
Clear, twisted protein tendrils extend from the surface of a blue bacterial cell. The protein tendrils become coated by gold nanoparticles, which start to glow red after they are heated. The heat causes the protein tendrils to break and the bacterial cell to burst.
Normally months or years of experiments would be required to test the interactions in every relevant environment. The researchers hope that their AI tool will speed up nanoparticle engineering by reducing trial and error.
“We hope to offer the scientific community a tool to produce nanoparticles on demand and obtain artificially manufactured materials with properties never seen before,” said Luis Liz-Marzán, a professor at CIC biomaGUNE and the University of Vigo and a co-principal investigator.

Beyond medicine, combinations of nanoparticles and proteins could be used to build devices that can guide and change light signals, enabling faster data transfer between nearby users within sight—such as between cars in a smart, interconnected fleet.
To build the AI-powered nanoparticle generator, the researchers will first make a training dataset using AlphaFold, an AI tool that predicts protein shapes. With the initial data, the researcher’s model will predict protein shapes that fit together like a key in a lock and how nanoparticles could be designed with similar shapes.
The researchers will verify model predictions by creating those matching nanoparticles and testing how the nanoparticles interact with their target proteins in lab experiments. Differences between the experimental results and the model’s predictions will then be used to refine the model.
The team will also need a training dataset of 3D images of their nanoparticles, which are conventionally made with time-consuming transmission electron tomography. It can sometimes take a full day to collect all of the images required for a 3D model of a single nanoparticle. Because of variations among the nanoparticles in any single batch, hundreds of 3D models need to be made.
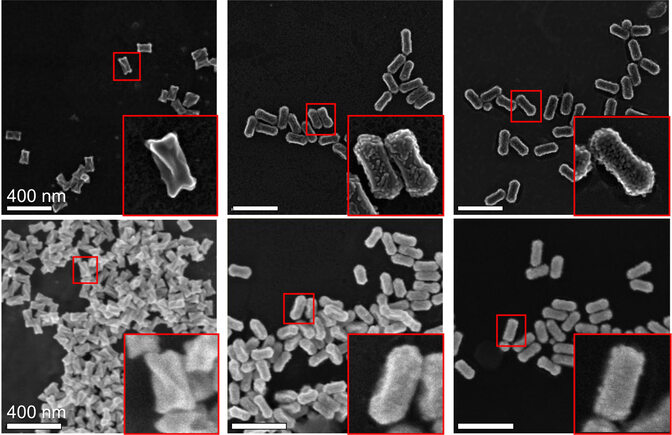
Sara Bals, a professor at the University of Antwerp’s EMAT electron microscopy center and a co-principal investigator, argues that nano researchers have been missing a trick: When the microscope’s electron beam passes through an object, electrons in the object’s surface escape, leaving behind holes. Other electrons will move to fill those holes, generating an electrical current shaped by the object’s topography. That current can be translated into a 3D image with a single snapshot.
A vertical, green electron beam sweeps over gold particles that have been strewn across a metal surface. As the beam moves over the particles, a high-contrast, gray-scale photo of the nanoparticles appears on screen.
“These interactions have always been there, but nobody has thought to use them for creating 3D images of nanoparticles,” Bals said. “With this technique, we can investigate hundreds of nanoparticles in a single image.”
Kotov is also the Joseph B. and Florence V. Cejka Professor of Engineering and a professor of macromolecular science and engineering and U-M’s Biointerfaces Institute.